Protein Family Sorter Service¶
Overview¶
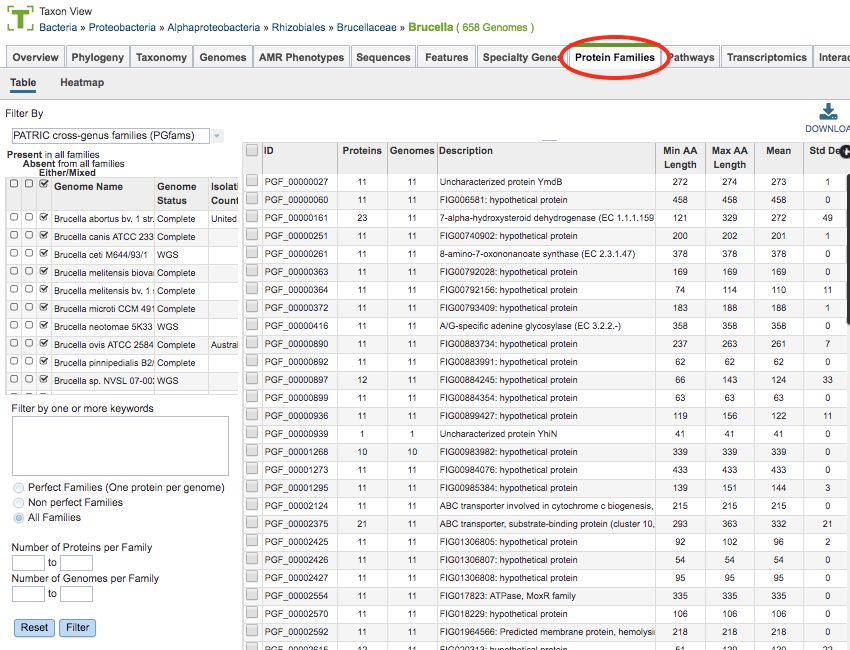
The bacterial Protein Family Sorter Service tool enables researchers to examine the distribution of protein families across a set of user-selected genomes. Results are displayed in a page showing all the families associated with the selected genomes, plus filter controls and an interactive heatmap.
See also¶
Using the Protein Family Sorter Service¶
The Protein Family Sorter submenu option under the Services main menu (Protein Tools category) opens the Protein Family Sorter input form (shown below).
Options¶
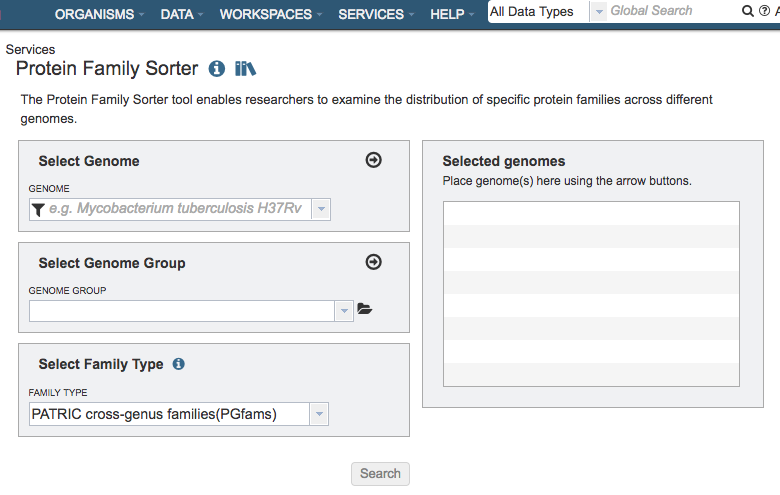
Select Genome¶
Input box for selecting genomes to include in the comparison. Click the arrow icon (->) to add to the set of Selected Genomes.
Select Genome Group¶
Input box for selecting genome group(s) to include in the comparison. Click the folder icon to navigate through the workspace to locate the genome group(s) of interest. Click the arrow icon (->) to add to the set of Selected Genomes. Note: You must be logged into BV-BRC to access genome groups in your private workspace.
Selected Genomes¶
List of genomes or genome groups that will be used in the comparison
Select Family Type¶
Family Type¶
Dropdown list of options of protein family types to use in the comparison:
Cenus-specific families(PLfams): The genus-specific protein families are computed using only proteins within a genus and more stringent criteria (MCL inflation = 3.0). This provides higher sequence similarity and better specificity while performing within-genus/species or close strain comparisons.
Cross-genus families(PGfams): The cross-genera protein families are computed by clustering representative proteins from the genus-specific families with slightly relaxed criteria (MCL inflation = 1.1). This allows cross-genera or distant homologs to cluster together, which is necessary to support cross-genera comparative analysis across all microbial genomes.
FIGFams: FIGfams are sets of isofunctional homologs, i.e. set of rotein sequences that are similar along their full length and believed to implement the same function. FIGfams are derived from a collection of functional subsystems, as well as correspondences between genes in closely related strains.
Output Results¶
Results from the service will be shown in a Protein Families Filter Tool and Table, shown below. See Protein Families Tab for a detailed description of how to use the Protein Family Sorter filters and heatmap.